Snap bean (Phaseolus vulgaris L.) is a significant vegetable crop globally, with its immature pods harvested as the edible part. Pod dimensions–length, width, and thickness–affect the size and ease of harvest. This is particularly for manual plucking, which is prevalent in China. Larger pods are often preferred for manual harvesting. Identifying the specific genes associated with pod width, thickness, and cross-sectional variation can be utilized in breeding programs to develop snap bean varieties with improved pod harvest and higher yields.
Recently, a research team led by Prof. XIA Zhengjun and Prof. LI Yanhua from the Key Laboratory of Black Soils Conservation and Utilization, Northeast Institute of Geography and Agroecology, Chinese Academy of Science (CAS) characterized and fine mapped the PvPW1 gene associated with pod width in snap bean.
In this study, a bi-parental F1 mapping population was developed from a cross between PI60234, a wide flattened pod cultivar, and Dalong1, a narrow oval pod cultivar was developed (Figure 1a). Through genotyping and phenotyping of recombinant plants, Phvul.006G072800, encoding the β-1,3-glucanase 9 protein, was identified as the causal gene for PvPW1, a quantitative trait locus associated with pod width in snap beans.
The results of an association analysis between pod width phenotype and the PvPW1G3555C genotype across 17 bi-parental F2 populations showed that the PvPW1G3555 allele was positively regulate pod width. 97.7% of the 133 wide pod accessions carriedPvPW1G3555(Figure 1b), while 82.1% of the 78 narrow pod accessions carriedPvPW1C3555(Figure 1c), indicating strong selection pressure onPvPW1during common bean breeding.
Furthermore, re-sequencing data from 59 common bean cultivars identified an 8-bp deletion in the intron linked to PvPW1C3555, leading to the development of the InDel marker of PvM436. Genotyping 317 common bean accessions with PvM436 demonstrated that accessions with PvM436247 and PvM436227 alleles had wider pods compared to those with PvM436219 allele (Figure 1d), establishing PvM436 as a reliable marker for molecular breeding in snap beans.
These findings highlight PvPW1 as a critical gene regulating pod width and underscore the utility of PvM436 in marker-assisted selection for snap bean breeding.
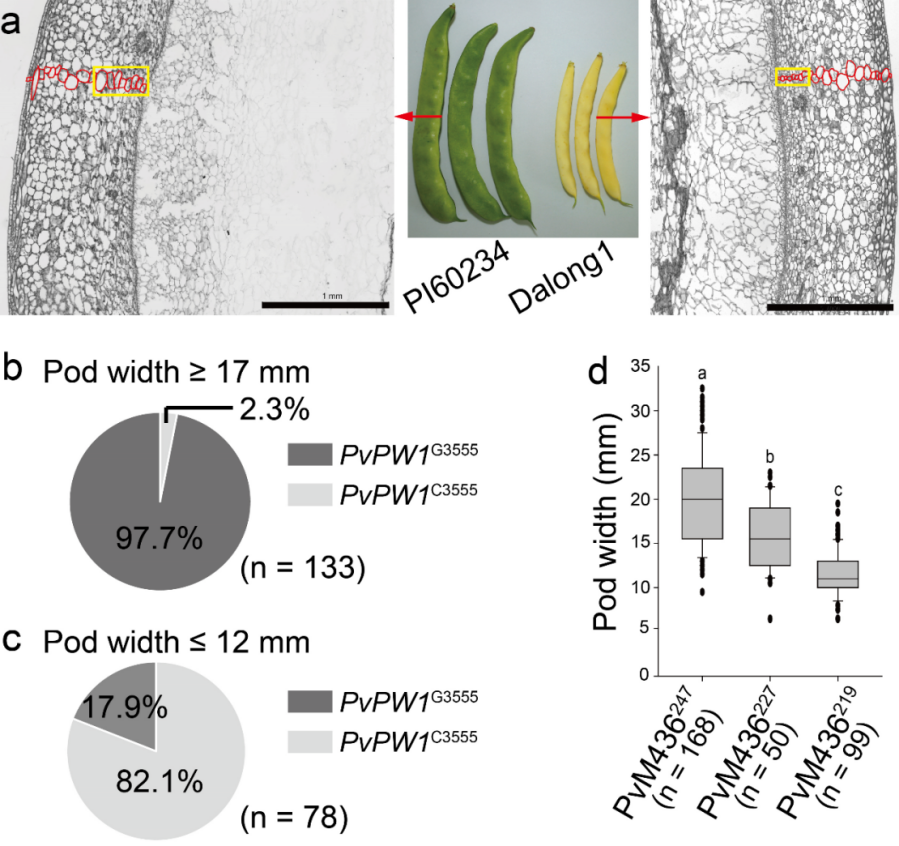
Figure 1a, The performance of pod width in PI60234 and Dalong1. Figure 1b, The frequency distribution of PvPW1G3555 and PvPW1C3555 genotypes in the 133 wide pod accessions. Figure 1c, The frequency distribution of PvPW1G3555 and PvPW1C3555 genotypes in the 78 narrow pod accessions. Figure 1d, Comparison of pod width of common bean accessions harboring PvM436247, PvM436227, and PvM436217.
Contact:
XU Kun
Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences
E-mail: xukun@iga.ac.cn
Reference link:
https://doi.org/10.1016/j.jgg.2024.09.020